Why Life Sciences AI Is a Search Problem (Part 2 of 5)

Photo by CHUTTERSNAP on Unsplash
The future of GenAI in pharma and healthcare isn’t about building bigger models — it’s about smarter retrieval.
By Harini Gopalakrishnan, Head of GTM, Vespa.ai
Originally presented at the Fierce Pharma Webinar: “You Have the Model, Now What? Lessons on Making AI Work in Life Sciences and Breaking the Chatbot Mirage.” This is a five part quick read series that summarizes the panel discussion highlighting key topics of the conversations.
This recap of the Fierce Pharma x Vespa.ai panel distills how leaders from Novo Nordisk, Alkermes, and Harvard Medical School are reframing AI as a search and retrieval problem, powered by context, tensors, and explainability.
→ Full video: fierce-pharma-webinar
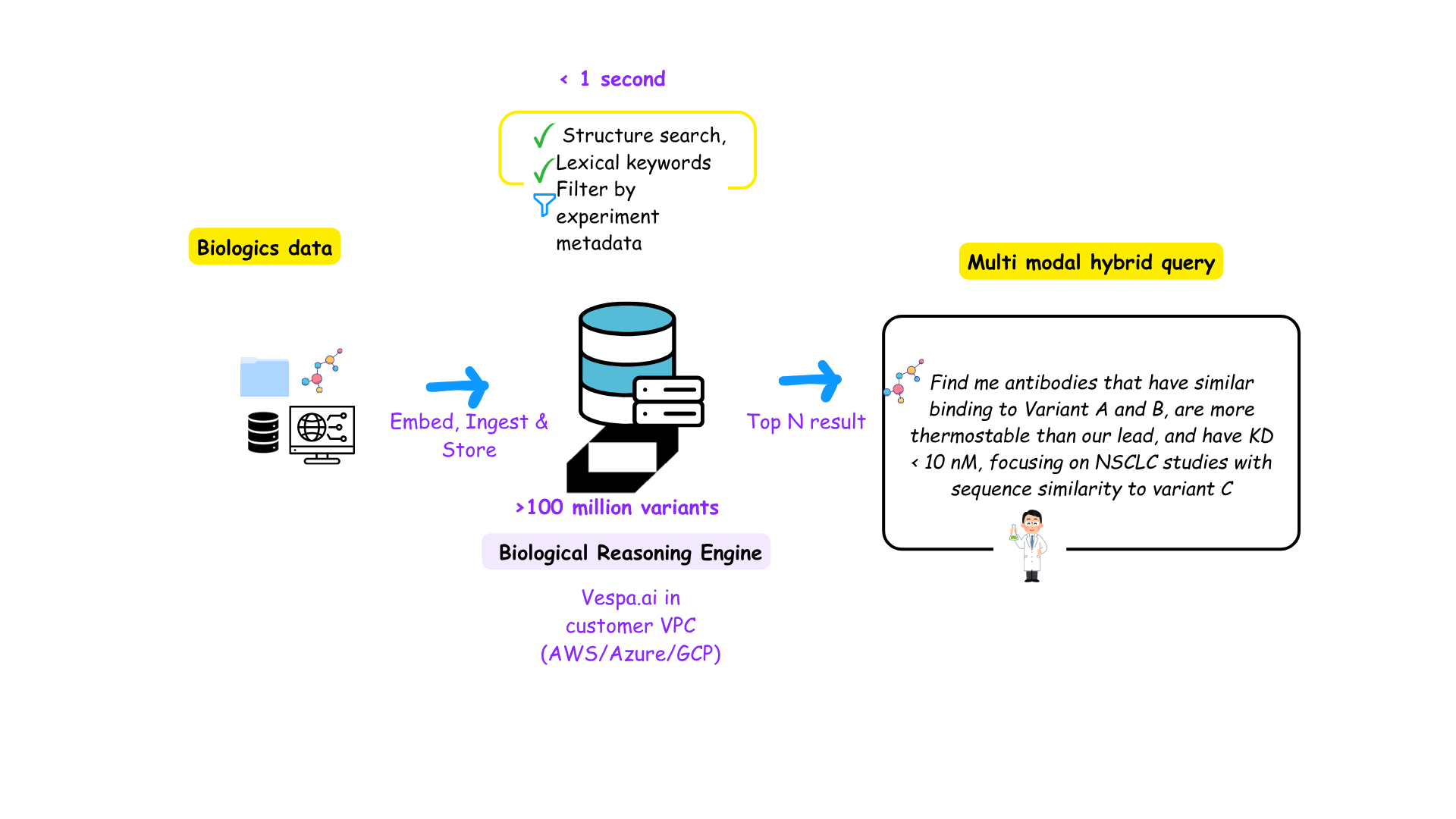
Pharma R&D: Drug Discovery as a Search Challenge
The focus now shifts to R&D in Lifesciences. R&D where techbios have been at the forefront in the last years. The 2026 Nobel prize was awarded to the creation of Alphafold in aiding protein folding.
With molecule and protein spaces spanning 10⁴⁰ to 10⁷⁰ possible combinations, drug discovery becomes an exercise in intelligent retrieval — sifting through an astronomical search space that’s literally larger than the number of atoms in the known universe.
The question was: How do (R&D) they see it as a Search problem? Are there any similarities between retrieval-first companies like Yahoo and Spotify and these protein language models?
Mark Brenkel, Head of Enterprise AI at Alkermes, described drug discovery as “finding a needle in a haystack larger than the known universe.”
“I think ‘find the needle in the haystack’ a great metaphor and it’s a good mental model for folks to have, like Internet search has figured out how to take really big spaces and make them searchable really fast.”
The process begins by constraining the search space using the intrinsic properties of molecules — for example, identifying whether a protein will fold into a biologically plausible structure. This reduces the candidate pool from 10⁴⁰ possibilities to something closer to 10⁸ or 10⁹ — still vast, but computationally tractable. Modern approaches use high-dimensional embeddings (tensors) to represent complex chemical, biological, and textual relationships in a unified mathematical form. (Read more: what is a tensor)
From there, external context such as assay results, publications, and prior experiments further refine the search. The relationships between these signals are captured within tensors, effectively encoding semantic and structural correlations.
“We’ve shifted from building bigger knowledge graphs to building smarter search spaces — using tensor embeddings to represent relationships between molecules, proteins, and diseases.”
Building a graph dynamically: the new paradigm
While traditional knowledge graphs required engineers to predefine entities and manually link relationships, tensorization allows these relationships to emerge dynamically. Models can now infer connections on demand and construct prompt-driven, dynamic graphs that not only retrieve relevant molecules but also feed downstream prediction and classification pipelines — accelerating discovery while preserving explainability.
“The summary of that is there is the modality, there is an embedding and there is a semantic relationship in that order. You need to create this, set this up before you frame it as a search problem and then you’re searching through the space”
Key Takeaways
-
Drug discovery = retrieval problem, not model problem.
-
Use embeddings to unify publications, assays, and molecular data.
-
Reduce over-engineering: let algorithms/models infer relationships across modalities.